Loading in Data
This is a quick tutorial to get you started with loading in your own data. feedr includes several wrapper functions that can be used to load and format your data.
A note about file structure
It’s important to remember that when specifying file locations, you must either specify a complete file location (e.g. “/home/steffi/Desktop/data.csv”) or an appropriate relative file location. A relative file location would be something like: “./Data/data.csv” which points to a file called data.csv which is in a folder called “Data”. The “./” indicates that “Data” is in the current working directory. If we used “../” that would indicate that “Data” was one directory up.
Also, remember that file locations are relative to where R’s working directory is, and this is not necessarily the same place as the R script with which you are working.
If you are using RStudio, it is highly recommended that you specify an RStudio project in the directory which holds your scripts. This way, anytime you open the file, the working directly is automatically set to your script directly.
This tutorial assumes that your data is stored in a folder called “Data” which is in turn stored in your R scripts folder.
load_raw()
This loads and formats a raw data file downloaded directly from RFID loggers setup in the same manner as the Thompson Rivers University loggers.
In the raw form, this data looks like:
GR10DATA
06200004BF 01/11/16 10:48:49
062000038F 01/11/16 10:48:50
06200004E4 01/11/16 10:48:55
06200004BF 01/11/16 10:50:02
06200004E4 01/11/16 10:51:07When we use load_raw() to import it, we get this:
r1 <- load_raw("./Data/Raw/exp2/GR10DATA_2016_01_16.TXT")## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_16.TXT...head(r1)## animal_id date time logger_id
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 GR10DATA
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 GR10DATA
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 GR10DATA
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 GR10DATA
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 GR10DATA
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 GR10DATANote that the logger_id has been extracted from the first line of the file. This is done by matching an expected pattern against the the contents of the first line (these patterns are called ‘Regular Expressions’).
The default pattern matches GR or GPR followed by 1 or 2 digits. If you need to specify a different pattern you can do so. For example, if your loggers were labeled “Logger_10” or “Logger_01”:
Logger_10
06200004BF 01/11/16 10:48:49
062000038F 01/11/16 10:48:50
06200004E4 01/11/16 10:48:55
06200004BF 01/11/16 10:50:02
06200004E4 01/11/16 10:51:07r1 <- load_raw("./Data/Raw/exp2/Logger_10_2016_01_16.TXT", logger_pattern = "Logger_[0-9]{2}")## Loading file ./Data/Raw/exp2/Logger_10_2016_01_16.TXT...head(r1)## animal_id date time logger_id
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 Logger_10
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 Logger_10
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 Logger_10
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 Logger_10
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 Logger_10
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 Logger_10Details
Alternatively, you can determine where load_raw() gets extra details from by specifying the details argument (defaults to details = 1).
For example:
1. details = 0
logger_id is in the file name, defined by the pattern logger_pattern (If logger_id is also the first line, need to skip that first line).
For example, if the file looks like:
Logger_21
06200004BF 01/11/16 10:48:49
062000038F 01/11/16 10:48:50
06200004E4 01/11/16 10:48:55
06200004BF 01/11/16 10:50:02
06200004E4 01/11/16 10:51:07Then this pattern won’t work because it matches the first line, not the file name:
load_raw("./Data/Raw/exp2/Logger_Data.TXT", logger_pattern = "Logger_[0-9]{2}", details = 0, skip = 1)## Loading file ./Data/Raw/exp2/Logger_Data.TXT...## Error: logger_id not detected in file nameBut this pattern will work:
load_raw("./Data/Raw/exp2/Logger_Data.TXT", logger_pattern = "Logger", details = 0, skip = 1)## Loading file ./Data/Raw/exp2/Logger_Data.TXT...## animal_id date time logger_id
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 Logger
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 Logger
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 Logger
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 Logger
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 Logger
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 Logger2. details = 1
logger_id is in the first line of the file, also defined by the pattern logger_pattern
load_raw("./Data/Raw/exp2/Logger_Data.TXT", logger_pattern = "Logger_[0-9]{2}", details = 1)## Loading file ./Data/Raw/exp2/Logger_Data.TXT...## animal_id date time logger_id
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 Logger_21
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 Logger_21
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 Logger_21
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 Logger_21
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 Logger_21
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 Logger_213. details = 2
logger_id is in the first line of the file, and lat/lon information is on the second line, in the format of “latitude, longitude” both in decimal format (spacing doesn’t matter, but the comma does):
Such a file would look like this:
Logger_21
53.89086,-122.81933
06200004BF 01/11/16 10:48:49
062000038F 01/11/16 10:48:50
06200004E4 01/11/16 10:48:55
06200004BF 01/11/16 10:50:02And would be loaded by load_raw() like this:
load_raw("./Data/Raw/exp2/Logger_Data.TXT", logger_pattern = "Logger_[0-9]{2}", details = 2)## Loading file ./Data/Raw/exp2/Logger_Data.TXT...## animal_id date time logger_id lat lon
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 Logger_21 53.89086 -122.8193
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 Logger_21 53.89086 -122.8193
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 Logger_21 53.89086 -122.8193
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 Logger_21 53.89086 -122.8193
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 Logger_21 53.89086 -122.8193
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 Logger_21 53.89086 -122.8193For more information on how to write Regular Expression patterns, see documentation for “Regular Expressions” (e.g. http://www.regular-expressions.info/tutorial.html)
load_raw_all()
The function load_raw_all() is a wrapper function which will automatically load and combine data contained in several different files in a single folder, or in a nested series of folders. Other files can be present, but all data files must be identifiable by a pattern (pattern argument) in the file name.
In this example our data files are stored in a folder called raw and there are several sets of data, each corresponding to an individual experiment which are then stored in their own folder called exp1, exp2, etc. Logger data files are identifiable by the characters ‘DATA’ present in the name (as in the above example), which is the default pattern:
r <- load_raw_all(r_dir = "./Data/Raw")## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_18.TXT...## Empty file skipped: ./Data/Raw/exp2/GR10DATA_2016_01_18.TXT## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_18.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_20.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp2/GR12DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR12DATA_2016_01_18.TXT...## Loading file ./Data/Raw/exp2/GR12DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_18.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_20.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_01.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_16.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_10.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_16.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_01.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_10.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_16.TXT...## Loading file ./Data/Raw/exp3/GR13DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR13DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR13DATA_2016_02_16.TXT...head(r)## animal_id date time logger_id
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 GR10DATA
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 GR10DATA
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 GR10DATA
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 GR10DATA
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 GR10DATA
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 GR10DATAsummary(r)## animal_id date time logger_id
## 062000038D: 4374 Min. :2016-01-06 Min. :2016-01-06 13:18:44 GR10DATA:5563
## 062000038F: 2750 1st Qu.:2016-01-14 1st Qu.:2016-01-14 13:16:54 GR11DATA:8697
## 0700EE2B10: 2614 Median :2016-01-21 Median :2016-01-21 12:12:26 GR12DATA:5649
## 062000014F: 2138 Mean :2016-01-25 Mean :2016-01-25 17:32:23 GR13DATA:8819
## 0700ED9E0E: 1809 3rd Qu.:2016-02-05 3rd Qu.:2016-02-05 09:11:10
## 0700EE0E42: 1675 Max. :2016-02-16 Max. :2016-02-16 14:30:29
## (Other) :13368(Note that empty files are skipped, but identified)
If your logger files don’t have an identifiable label, but are the only csv files in the folders, you could use:
r <- load_raw_all(r_dir = "./Data/Raw", pattern = ".csv")Extra details
In this example we have several different experiments, which we’ll probably want to identify in our data. This is where the extra_ arguments come in.
list.files("./Data/Raw")## [1] "exp2" "exp3"In our example, each experiment is stored in its own folder (‘exp2’ and ‘exp3’). Therefore we can tell our function to identify patterns (extra_pattern) in the file names and store the values in an extra column (extra_name):
r <- load_raw_all(r_dir = "./Data/Raw", extra_pattern = "exp[2-3]{1}", extra_name = "experiment")## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_18.TXT...## Empty file skipped: ./Data/Raw/exp2/GR10DATA_2016_01_18.TXT## Loading file ./Data/Raw/exp2/GR10DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_18.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_20.TXT...## Loading file ./Data/Raw/exp2/GR11DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp2/GR12DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR12DATA_2016_01_18.TXT...## Loading file ./Data/Raw/exp2/GR12DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_16.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_18.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_20.TXT...## Loading file ./Data/Raw/exp2/GR13DATA_2016_01_25.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_01.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR10DATA_2016_02_16.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_10.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR11DATA_2016_02_16.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_01.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_10.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR12DATA_2016_02_16.TXT...## Loading file ./Data/Raw/exp3/GR13DATA_2016_02_06.TXT...## Loading file ./Data/Raw/exp3/GR13DATA_2016_02_13.TXT...## Loading file ./Data/Raw/exp3/GR13DATA_2016_02_16.TXT...head(r)## animal_id date time logger_id experiment
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 GR10DATA exp2
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 GR10DATA exp2
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 GR10DATA exp2
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 GR10DATA exp2
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 GR10DATA exp2
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 GR10DATA exp2“exp[1-2]{1}” matches the exact characters “exp” followed by either a 1 or a 2 of which there is exactly 1. The values matching this pattern are then stored in a new column called ‘experiment’.
Because here the loggers were RFID-enabled feeders reused for different experiments, some logger have the same id, but a different lat/lon. However, logger_ids need to be unique or we will have problems later on, so let’s create unique logger_id names:
r$logger_id <- paste(r$experiment, r$logger_id, sep = "-")
head(r)## animal_id date time logger_id experiment
## 1 06200004BF 2016-01-11 2016-01-11 10:48:49 exp2-GR10DATA exp2
## 2 062000038F 2016-01-11 2016-01-11 10:48:50 exp2-GR10DATA exp2
## 3 06200004E4 2016-01-11 2016-01-11 10:48:55 exp2-GR10DATA exp2
## 4 06200004BF 2016-01-11 2016-01-11 10:50:02 exp2-GR10DATA exp2
## 5 06200004E4 2016-01-11 2016-01-11 10:51:07 exp2-GR10DATA exp2
## 6 06200004BE 2016-01-11 2016-01-11 10:51:51 exp2-GR10DATA exp2Logger details
Because raw logger data doesn’t include logger specific details, we should probably include some extra data for use later (visualizations, etc.):
We can do this the same way as we did above (see ‘Details’ under load_raw()), or by merging our raw data with a logger index file. This method works best when you have multiple details you’d like to add (more than just lat/lon information), or when your raw files don’t contain lat/lon information already.
## Open logger index
l_index <- read.csv("./Data/chickadees_logger_index.csv")
head(l_index)## experiment logger_name lat lon
## 1 exp2 GR10DATA 53.89086 -122.8193
## 2 exp2 GR11DATA 53.88999 -122.8210
## 3 exp2 GR12DATA 53.88997 -122.8193
## 4 exp2 GR13DATA 53.89088 -122.8208
## 5 exp3 GR10DATA 53.88763 -122.8217
## 6 exp3 GR11DATA 53.88821 -122.8205l_index$logger_id <- paste(l_index$experiment, l_index$logger_name, sep = "-")
## Merge logger index into RFID data, matching 'experiment' and 'logger_id'
r <- merge(r, l_index, by = c("experiment", "logger_id"))
head(r)## experiment logger_id animal_id date time logger_name lat lon
## 1 exp2 exp2-GR10DATA 06200004BF 2016-01-11 2016-01-11 10:48:49 GR10DATA 53.89086 -122.8193
## 2 exp2 exp2-GR10DATA 062000038F 2016-01-11 2016-01-11 10:48:50 GR10DATA 53.89086 -122.8193
## 3 exp2 exp2-GR10DATA 06200004E4 2016-01-11 2016-01-11 10:48:55 GR10DATA 53.89086 -122.8193
## 4 exp2 exp2-GR10DATA 06200004BF 2016-01-11 2016-01-11 10:50:02 GR10DATA 53.89086 -122.8193
## 5 exp2 exp2-GR10DATA 06200004E4 2016-01-11 2016-01-11 10:51:07 GR10DATA 53.89086 -122.8193
## 6 exp2 exp2-GR10DATA 06200004BE 2016-01-11 2016-01-11 10:51:51 GR10DATA 53.89086 -122.8193This data is now ready for housekeeping or go straight to transformations!
ui_import()
ui_import() is a helper function that launches a stand-alone shiny app to give you a user-interface for importing data.
library(feedrUI)
my_imported_data <- ui_import()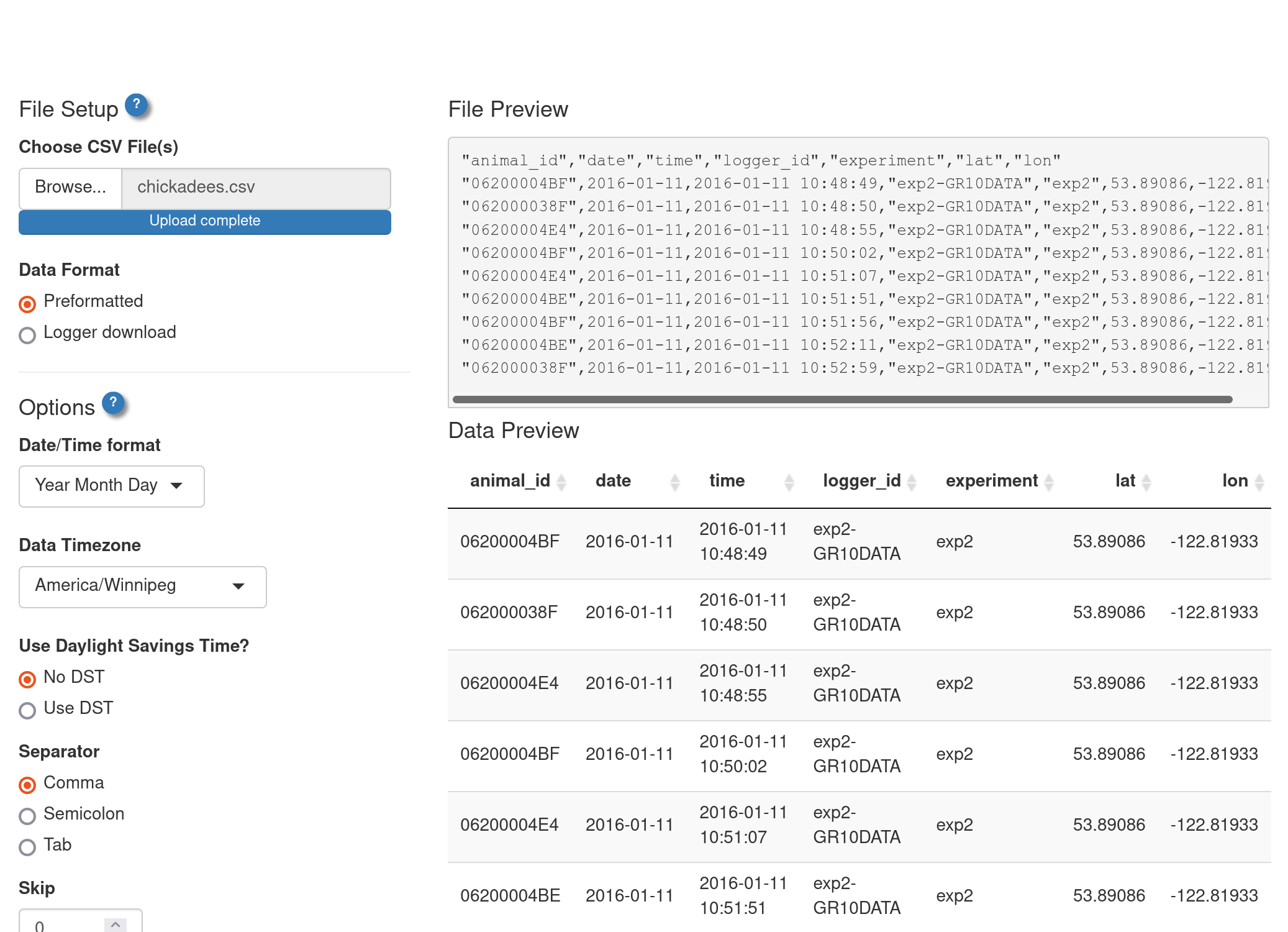
Back to top
Go back to home | Continue with housekeeping